Practice Corner MCQ
Quiz Summary
0 of 79 Questions completed
Questions:
Information
You have already completed the quiz before. Hence you can not start it again.
Quiz is loading…
You must sign in or sign up to start the quiz.
You must first complete the following:
Results
Results
0 of 79 Questions answered correctly
Your time:
Time has elapsed
You have reached 0 of 0 point(s), (0)
Earned Point(s): 0 of 0, (0)
0 Essay(s) Pending (Possible Point(s): 0)
Categories
- Not categorized 0%
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
- 53
- 54
- 55
- 56
- 57
- 58
- 59
- 60
- 61
- 62
- 63
- 64
- 65
- 66
- 67
- 68
- 69
- 70
- 71
- 72
- 73
- 74
- 75
- 76
- 77
- 78
- 79
- Current
- Review
- Answered
- Correct
- Incorrect
-
Question 1 of 79
1. Question
\(
\text { What does } \mathrm{A} \text { and } \mathrm{B} \text { represent in the given representation? }
\)
Sugar+Nitrogenous base =A
Phosphate group+sugar+nitrogenous base =BCorrectIncorrectHint
(c)
-
Question 2 of 79
2. Question
In DNA strand, the nucleotides are linked together by
CorrectIncorrectHint
(b) : Two nucleotides are linked through \(3^{\prime} \rightarrow 5^{\prime}\) phosphodiester linkage to form a dinucleotide. More nucleotides are joined in a similar manner to form a polynucleotide chain.
-
Question 3 of 79
3. Question
In a DNA molecule, the phosphate group is attached to carbon ____ of the sugar residue of its own nucleotide and carbon ____ of the sugar residue of the next nucleotide by _____ bonds.
CorrectIncorrectHint
(a)
-
Question 4 of 79
4. Question
DNA as an acidic substance present in nucleus was first identified by _____ in 1869; he named it as _____.
CorrectIncorrectHint
(a)
-
Question 5 of 79
5. Question
Which of the following pairs is incorrectly matched?
CorrectIncorrectHint
(d) : DNA (deoxyribonucleic acid) is an acidic biomolecule.
-
Question 6 of 79
6. Question
Watson and Crick (1953) proposed DNA double helix model and won the Nobel Prize; their model of DNA was based on
(i) X-ray diffraction studies of DNA done by Wilkins and Franklin
(ii) Chargaff’s base equivalence rule
(iii) Griffith’s transformation experiment
(iv) Meselson and Stahl’s experiment.CorrectIncorrectHint
(b) : In 1953, James Watson and Francis Crick based on the X-ray diffraction data produced by Maurice Wilkins and Rosalind Franklin, proposed double helix model for the structure of DNA. One of the hallmarks of their proposition was base pairing between the two strands of polynucleotide chains. However, this proposition was also based on the observation of Erwin Chargaff (1950) that for a double stranded DNA, the ratios between adenine and thymine, and quanine and cytosine are constant and equats one.
-
Question 7 of 79
7. Question
If a double stranded DNA has \(20 \%\) of cytosine, what will be the percentage of adenine in it?
CorrectIncorrectHint
(c) : According to Chargaff’s rule, the amount of adenine is always equal to that of thymine and the amount of guanine is always equal to that of cytosine, i.e., \(A=T\) and \(G=C\). Also, the purines and pyrimidines are always in equal amounts, i.e., \(\mathrm{A}+\mathrm{G}\) \(=\mathrm{T}+\mathrm{C}\). Now, given dsDNA has \(20 \%\) cytosine and hence guanine will be also \(20 \%\).
So, \(A+\) I must be \(60 \%\). Therefore, percentage of adenine would be \(60 / 2=30 \%\). -
Question 8 of 79
8. Question
In one polynucleotide strand of a DNA molecule, the ratio of \(\mathrm{A}+\mathrm{T} / \mathrm{G}+\mathrm{C}\) is 0.3 . What is the \(\mathrm{A}+\mathrm{G} / \mathrm{T}+\mathrm{C}\) ratio of the entire DNA molecule?
CorrectIncorrectHint
(d): According to Chargaff’s rules (1950), purine and pyrimidine base pairs are present in equal amounts in a DNA molecule.
-
Question 9 of 79
9. Question
If the sequence of bases in one strand of DNA is ATGCATGCA, what would be the sequence of bases on complementary strand?
CorrectIncorrectHint
(c) : Adenine always pairs with thymine and cytosine always pairs with guanine. Hence, sequence of bases on complementary strand would be TACGTACGT.
-
Question 10 of 79
10. Question
Refer to the given figure.
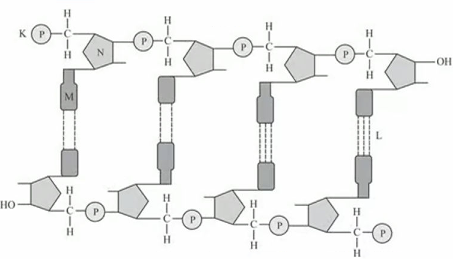
Select the option which correctly identifies K,L M and N.
\(
\begin{array}{|l|l|l|l|l|}
\hline & \text { K } & \text { L } & \text { M } & \text { N } \\
\hline \text { (a) } & 5^{\prime} \text { end } & \text { Phosphodiester bond } & \text { Thymine } & \text { Ribose sugar } \\
\hline \text { (b) } & 3^{\prime} \text { end } & \text { Glycosidic bond } & \text { Guanine } & \text { Deoxyribose sugar } \\
\hline \text { (c) } & 5^{\prime} \text { end } & \text { Hydrogen bond } & \text { Adenine } & \text { Ribose sugar } \\
\hline \text { (d) } & 5^{\prime} \text { end } & \text { Hydrogen bond } & \text { Adenine } & \text { Deoxyribose sugar } \\
\hline
\end{array}
\)CorrectIncorrectHint
(d)
-
Question 11 of 79
11. Question
The given flow chart represents the flow of genetic information between biomolecules. Identify the processes A, B, C and D and select the correct option.
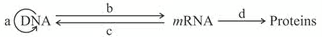 CorrectIncorrect
CorrectIncorrectHint
(c) : DNA replication refers to the process of formation of two similar copies of DNA using parental DNA as template. The expression of the genetic material occurs normally through the production of proteins. This involves two consecutive steps. These are transcription and translation. The DNA codes for the production of messenger RNA (mRNA) during transcription. mRNA carries coded information to ribosomes. The ribosomes read this information and use it for protein synthesis. This process is called translation. Crick described this unidirectional flow of information in 1958 as the central dogma of molecular biology. Many tumour viruses contain RNA as genetic material and replicate by first synthesising a complementary DNA. This process is called reverse transcription (teminism). It is carried out by an RNA-dependent DNA polymerase called reverse transcriptase.
-
Question 12 of 79
12. Question
Synthesis of DNA from RNA is explained by
CorrectIncorrectHint
(d) : DNA replication refers to the process of formation of two similar copies of DNA using parental DNA as template. The expression of the genetic material occurs normally through the production of proteins. This involves two consecutive steps. These are transcription and translation. The DNA codes for the production of messenger RNA (mRNA) during transcription. mRNA carries coded information to ribosomes. The ribosomes read this information and use it for protein synthesis. This process is called translation. Crick described this unidirectional flow of information in 1958 as the central dogma of molecular biology. Many tumour viruses contain RNA as genetic material and replicate by first synthesising a complementary DNA. This process is called reverse transcription (teminism). It is carried out by an RNA-dependent DNA polymerase called reverse transcriptase.
-
Question 13 of 79
13. Question
Histone proteins are
CorrectIncorrectHint
(b) : In eukaryotes, DNA organisation is complex. There is a set of positively charged, basic proteins called histones. Histones are rich in the basic amino acid residues lysine and arginine. There are five types of histone proteins – \(\mathrm{H}_1, \mathrm{H}_2 \mathrm{~A}, \mathrm{H}_2 B, \mathrm{H}_3\) and \(\mathrm{H}_4\). Four of them \(\left(\mathrm{H}_2 \mathrm{~A}, \mathrm{H}_2 \mathrm{~B}, \mathrm{H}_3\right.\) and \(\left.\mathrm{H}_4\right)\) produce histone octamer called nu body or core of nucleosome. The negatively charged DNA is wrapped around the positively charged histone octamer to form nucleosome. DNA connecting two adjacent nucleosomes is called linker DNA which bears \(\mathrm{H}_1\) histone proteins.
-
Question 14 of 79
14. Question
What does the given diagram represent?
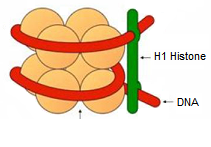 CorrectIncorrect
CorrectIncorrectHint
(a)
-
Question 15 of 79
15. Question
The structure in chromatin seen as ‘beads-on string’ when viewed under electron microscope are called
CorrectIncorrectHint
(d) : Nucleosomes constitute the repeating unit of a structure in nucleus called chromatin, thread-like stained (coloured) bodies seen in nucleus. The nucleosomes in chromatin are seen as ‘beadson-string’ structure when viewed under electron microscope.
-
Question 16 of 79
16. Question
Refer the given figure of nucleosome and select the option that correctly identifies the parts A, B and C .
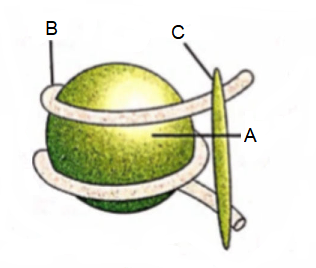 CorrectIncorrect
CorrectIncorrectHint
(c)
-
Question 17 of 79
17. Question
Read the following statements and select the correct option.
(i) Loosely packed and lightly stained region of chromatin are called as heterochromatin.
(ii) Densely packed and dark stained region of chromatin are called as euchromatin.
(iii) A typical nucleosome contains 200 bp of DNA helix.CorrectIncorrectHint
(b) : A typical nucleosome contains 200 bp of DNA helix. Nucleosomes constitute the repeating unit of a structure in nucleus called chromatin. Chromatin is held over a scaffold of non-histone chromosomal (or NHC ) proteins. At some places chromatin is densely packed to form darkly stained heterochromatin. At other places chromatin is loosely packed. It is called euchromatin. It is transcriptionally active chromatin whereas heterochromatin is transcriptionally inactive and late replicating or heteropycnotic.
-
Question 18 of 79
18. Question
Other than DNA polymerase, which of the following enzymes is involved in DNA synthesis?
CorrectIncorrectHint
(d) : Process of DNA synthesis whereby a parent DNA molecule is faithfully copied, giving rise to two identical daughter molecules is called DNA replication. in DNA synthesis, DNA polymerase plays an important role having the capability to elongate an existing DNA strand but cannot initiate the synthesis. So, the synthesis is initiated with the help of RNA primer formed by RNA primase. RNA primase synthesises the short RNA primer of about 10 nucleotides that is elongated by DNA polymerase to form an Okazaki fragment of DNA during DNA replication. Helicase unzips the two strands of DNA and topoisomerase reduces the coiling tension developed due to the unwinding of the two strands.
-
Question 19 of 79
19. Question
Refer to the given steps of DNA replication.
(i) Exposure of DNA strands
(ii) Synthesis of RNA primer
(iii) Activation of deoxyribonucleotides
(iv) Chain formation
(v) Base pairing
(vi) Proof reading and DNA repair
(vii) DNA polymerase attaches at Ori site
Select the correct sequence of DNA replication.CorrectIncorrectHint
(a)
-
Question 20 of 79
20. Question
Which of the following figures correctly represents the replication fork formed during DNA replication?
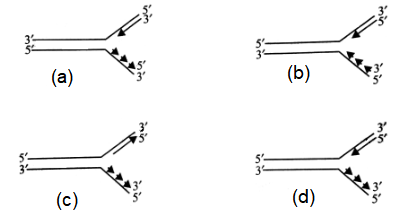 CorrectIncorrect
CorrectIncorrectHint
(d) : Synthesis of DNA by DNA polymerases occurs only in \(5^{\prime} \rightarrow 3^{\prime}\) direction. One strand called leading strand, is copied in the same direction as the unwinding helix. The other strand is known as lagging strand. Replication of lagging strand is in a discontinuous way, and in the direction of growth of lagging stand is \(3^{\prime} \rightarrow 5^{\prime}\) though in short segments of DNA which are alwavs in the \(5^{\prime} \rightarrow\) \(3^{\prime}\) direction. These short segments are called Okazaki fragments joined together by the action of DNA ligase.
-
Question 21 of 79
21. Question
Select the incorrect statement regarding DNA replication.
CorrectIncorrectHint
(d) : Synthesis of DNA by DNA polymerases occurs only in \(5^{\prime} \rightarrow 3^{\prime}\) direction. One strand called leading strand, is copied in the same direction as the unwinding helix. The other strand is known as lagging strand. Replication of lagging strand is in a discontinuous way, and in the direction of growth of lagging stand is \(3^{\prime} \rightarrow 5^{\prime}\) though in short segments of DNA which are alwavs in the \(5^{\prime} \rightarrow\) \(3^{\prime}\) direction. These short segments are called Okazaki fragments joined together by the action of DNA ligase.
-
Question 22 of 79
22. Question
Which of the following differences are incorrect between leading and lagging strands of DNA?
\(
\begin{array}{|c|c|c|}
\hline & \text { Leading strand } & \text { Lagging strand } \\
\hline \text { (i) } & \begin{array}{l}
\text { It does not require } \\
\text { DNA ligase for its } \\
\text { growth. }
\end{array} & \begin{array}{l}
\text { DNA ligase is required } \\
\text { for joining Okazaki } \\
\text { fragments. }
\end{array} \\
\hline \text { (ii) } & \begin{array}{l}
\text { Formation of leading }
\end{array} & \begin{array}{l}
\text { Formation of lagging } \\
\text { strand is quite rapid. }
\end{array} \\
\hline
\end{array}
\)\(
\begin{array}{|c|c|c|}
\hline \text { (iii) } & \begin{array}{l}
\text { Its template opens in } \\
5^{\prime} \rightarrow 3^{\prime} \text { direction. }
\end{array} & \begin{array}{l}
\text { Its template opens in } \\
3^{\prime} \rightarrow 5^{\prime} \text { direction. }
\end{array} \\
\hline \text { (iv) } & \begin{array}{l}
\text { Formation of leading } \\
\text { strand begins } \\
\text { immediately at } \\
\text { the beginning of }
\end{array} & \begin{array}{l}
\text { Formation of lagging } \\
\text { strand begins a bit later } \\
\text { than that of leading } \\
\text { strand. }
\end{array} \\
\hline
\end{array}
\)CorrectIncorrectHint
(c)
-
Question 23 of 79
23. Question
DNA replication takes place at _____ phase of the cell cycle.
CorrectIncorrectHint
(b) : In eukaryotes, the replication of DNA takes place at S-phase of the cell cycle. The replication of DNA and cell division cycle should be coordinated. A failure in cell division after DNA replication results into chromosomal anomaly.
-
Question 24 of 79
24. Question
The process of copying genetic information from one strand of DNA into RNA is termed as ______.
CorrectIncorrectHint
(b) : The process of copying genetic information from one strand of the DNA into RNA is termed as transcription. Unlike the process of replication, which once sets in, the total DNA of an organism gets duplicated, in transcription only a segment of DNA and only one of the strands is copied into RNA.
-
Question 25 of 79
25. Question
The given flow chart shows central dogma reverse

Enzymes used in processes A, B and C are respectively
CorrectIncorrectHint
(b)
-
Question 26 of 79
26. Question
The enzyme DNA dependent RNA polymerase catalyses the polymerisation reaction in _____ direction.
CorrectIncorrectHint
(a) : The two strands have opposite polarity and the DNA-dependent RNA polymerase also catalyse the polymerisation in only one direction, that is, \(5^{\prime} \rightarrow 3^{\prime}\), the strand that has the polarity \(3^{\prime} \rightarrow 5^{\prime}\) acts as a template, and is also referred to as template strand. The other strand which has the polarity \(5^{\prime} \rightarrow 3^{\prime}\) and the sequence same as RNA (except thymine at the place of uracil) is displaced during transcription. This strand (which does not code for anything) is referred to as coding strand.
-
Question 27 of 79
27. Question
What would be the base sequence of RNA transcript obtained from the given DNA segment?
\(5^{\prime}\) – G CATT C G G CTA G TAA C \(-3^{\prime}\) Coding strand of DNA
\(3^{\prime}\) – C GTAAGCCGATCATTG – \(5^{\prime}\) Non-coding strand of DNACorrectIncorrectHint
(a)
-
Question 28 of 79
28. Question
If the sequence of bases in coding strand of DNA is ATTCGATG, then the sequence of bases in mRNA will be
CorrectIncorrectHint
(d) : mRNA strand is complementary to template strand of DNA, while coding strand has same sequence as RNA (except thymine at the place of uracil). Hence, the sequence of bases in mRNA will be AUUCGAUG.
-
Question 29 of 79
29. Question
If the sequence of bases in DNA is GCTTAGGCAA, then the sequence of bases in its transcript will be
CorrectIncorrectHint
(c) : mRNA strand is complementary to one of the DNA strands i.e., template strand. In RNA, uracil is present instead of thymine which is complementary to adenine. Cytosine and guanine are also complementary to each other. Hence, the sequence of bases in transcript would be CGAAUCCGUU
-
Question 30 of 79
30. Question
Given diagram represents the components of a transcription unit. Select the correct answer regarding it.
 CorrectIncorrect
CorrectIncorrectHint
(c)
-
Question 31 of 79
31. Question
Transcription unit
CorrectIncorrectHint
(c) : Transcription unit is the distance between sites of initiation and termination by RNA polymerase. It may include more than one gene. RNA polymerase produces transcription unit that extends from the promoter to the termination sequences.
-
Question 32 of 79
32. Question
During transcription, the site of DNA molecule at which RNA polymerase binds is called
CorrectIncorrectHint
(a) : The RNA polymerase enzyme binds to a specific site called promoter and initiates transcription. The promoter region determines which DNA strand is to be transcribed. Thus, a promoter region has RNA polymerase recognition site and RNA polymerase binding site.
-
Question 33 of 79
33. Question
Select the correct match of enzyme with its related function.
CorrectIncorrectHint
(d)
-
Question 34 of 79
34. Question
Polycistronic messenger RNA (mRNA) usually occurs in
CorrectIncorrectHint
(d) : When a particular gene codes for a mRNA strand it is said to be monocistronic or monogenic. When several genes (cistrons) are transcribed into a single mRNA molecule, it is described as polycistronic or polygenic. Prokaryotic mRNA are polycistronic.
-
Question 35 of 79
35. Question
The given figure represents the process of transcription in bacteria.
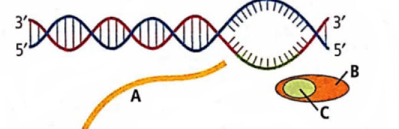
Select the option which correctly labels A, B and C.
CorrectIncorrectHint
(b)
-
Question 36 of 79
36. Question
Refer to the given diagram. What does it represent?
 CorrectIncorrect
CorrectIncorrectHint
(b) : The given diagram represents post-transcriptional processing resulting in the formation of mRNA. Since, introns and exons are present, it is transcription in eukaryotes.
-
Question 37 of 79
37. Question
In transcription in eukaryotes, heterogenous nuclear RNA (hnRNA) is transcribed by
CorrectIncorrectHint
(b) : Eukaryotes have three RNA polymerases. RNA polymerase 1 is located in the nucleolus and transcribes for rRNAs (285, 185 and 5.85 ) whereas RNA polymerase II is localised in the nucleoplasm and used for hnRNA, mRNA and RNA polymerase III is localised in the nucleus, possibly the nucleolarnucleoplasm interface and transcribes for \(\)t\(\) RNA, 55 rRNA and snRNAs.
-
Question 38 of 79
38. Question
Methyl guanosine triphosphate is added to the \(5^{\prime}\) end of hnRNA in a process of
CorrectIncorrectHint
(b) : At 5′ end of hnRNA, a cap is formed by modification of GTP into 7 -methyl guanosine or \(7^{\mathrm{M}} \mathrm{Gppp}\). This process is called capping and it protects the mRNA from degradation by nucleases.
-
Question 39 of 79
39. Question
Select the correct statements regarding the process of transcription in eukaryotes.
(i) The strand of dsDNA which takes part in transcription process is called as coding strand.
(ii) The enzyme RNA polymerase can catalyse polymerisation only in one direction i.e., \(5^{\prime} \rightarrow 3^{\prime}\).
(iii) An unusual nucleotide methyl guanosine triphosphate is added to the \(5^{\prime}\) end of hnRNA during capping.
(iv) During tailing process, adenylate residues (200300) are added at \(3^{\prime}\) end in a template independent manner.CorrectIncorrectHint
(c)
-
Question 40 of 79
40. Question
In eukaryotes, the process of processing of primary transcript involves
CorrectIncorrectHint
(d) : The primary mRNA transcript is longer and localised into the nucleus, where it is also called heterogenous nuclear RNA (hnRNA) or pre-mRNA. At the \(5^{\prime}\) end of hnRNA, a cap (consisting of 7 -methyl guanosine triphosphate or 7 mG ) and a tail of poly \(A\) (Adenylate residues) at the \(3^{\prime}\) end are added. These processes are respectively called as capping and tailing. The cap is a chemically modified molecule of guanosine triphosphate (GTP). The primary mRNAs are made up of two types of segments; non-coding introns and the coding exons. The introns are removed by a process called RNA splicing and the exons are joined in a defined order.
-
Question 41 of 79
41. Question
Match column I with column II and select the correct option from the given codes
\(
\begin{array}{|c|l|c|l|}
\hline & \text { Column I } & & \text { Column II } \\
\hline \text { A. } & \text { Sigma factor } & \text { (i) } & 5^{\prime}-3^{\prime} \\
\hline \text { B. } & \text { Capping } & \text { (ii) } & \text { Initiation } \\
\hline \text { C. } & \text { Tailing } & \text { (iii) } & \text { Termination } \\
\hline \text { D. } & \text { Coding strand } & \text { (iv) } & 5^{\prime} \text { end } \\
\hline & & \text { (v) } & 3^{\prime} \text { end } \\
\hline
\end{array}
\)CorrectIncorrectHint
(b)
-
Question 42 of 79
42. Question
Identify A, B, C and D in the given diagram of mRNA.
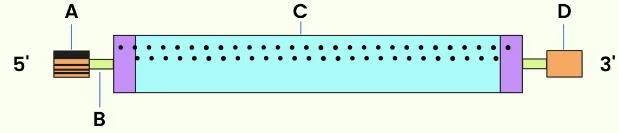 CorrectIncorrect
CorrectIncorrectHint
(c)
-
Question 43 of 79
43. Question
The fully processed hnRNA is called as (i) and is transported out of the (ii) into the (iii) for translation.
CorrectIncorrectHint
(a)
-
Question 44 of 79
44. Question
Refer to the given sequence of steps and select the correct option.
\(
\mathrm{DNA} \xrightarrow{(\text { i) }} \mathrm{hnRNA}\xrightarrow{(\text { ii) }} \text { mRNA } \xrightarrow{\text { (iii) }} \text { Proteins }
\)CorrectIncorrectHint
(c) : DNA contains genetic information which is transcribed via mRNA. mRNA is not made directly in eukaryotic cell. It is transcribed in the nucleus as hnRNA which contains introns and exons. The introns are removed by RNA splicing leaving behind exons which contain the information. The exonic regions of RNA are joined together to produce a single chain RNA required for functioning as translational template. This information is then translated into polypeptide chains.
-
Question 45 of 79
45. Question
The three codons which result in the termination of polypeptide chain synthesis are
CorrectIncorrectHint
(b)
-
Question 46 of 79
46. Question
Match column I with column II and select the correct option from the given codes.
\(
\begin{array}{|c|l|c|l|}
\hline & \begin{array}{l}
\text { Column I } \\
\text { (Codons) }
\end{array} & & \begin{array}{l}
\text { Column II } \\
\text { (Translated amino acid) }
\end{array} \\
\hline \text { A. } & \text { UUU } & \text { (i) } & \text { Serine } \\
\hline \text { B. } & \text { GGG } & \text { (ii) } & \text { Methionine } \\
\hline \text { C. } & \text { UCU } & \text { (iii) } & \text { Phenylalanine } \\
\hline \text { D. } & \text { CCC } & \text { (iv) } & \text { Glycine } \\
\hline \text { E. } & \text { AUG } & \text { (v) } & \text { Proline } \\
\hline
\end{array}
\)CorrectIncorrectHint
(a)
-
Question 47 of 79
47. Question
Amino acids which are specified by single codons are
CorrectIncorrectHint
(b) : Since there are 64 triplet codons and only 20 amino acids, the incorporation of some amino acids must be influenced by more than one codon. Onty triptophan (UGG) and methionine (AUG) are specified by single codons. All other amino acids are specified by two (e.g., phenylalanine – UUU, UUC) to six (e.g., arginine-CGU, CGC, CGA, CGG, AGA, AGG) codons. The latter are called degenerate or redundant codons. In degenerate codons, generally the first two nitrogen bases are similar while the third one is different. As the third nitrogen base has no effect on coding, the same is called wobble position.
-
Question 48 of 79
48. Question
Which out of the following statements is incorrect?
CorrectIncorrectHint
(a) : Since there are 64 triplet codons and only 20 amino acids, genetic code is non-ambiguous. Non-ambiguous code means that there is no ambiguity about a particular code. One codon specifies only one amino acid and not any other except GUG which normally codes for valine but in certain conditions it also codes for N -formyl methionine as initiation codon.
-
Question 49 of 79
49. Question
Some amino acids are coded by more than one codon, hence the genetic code is
CorrectIncorrectHint
(b) : Since there are 64 triplet codons and only 20 amino acids, the incorporation of some amino acids must be influenced by more than one codon. Onty triptophan (UGG) and methionine (AUG) are specified by single codons. All other amino acids are specified by two (e.g., phenylalanine – UUU, UUC) to six (e.g., arginine-CGU, CGC, CGA, CGG, AGA, AGG) codons. The latter are called degenerate or redundant codons. In degenerate codons, generally the first two nitrogen bases are similar while the third one is different. As the third nitrogen base has no effect on coding, the same is called wobble position.
-
Question 50 of 79
50. Question
Read the following statements.
(i) One codon codes for only one amino acid.
(ii) Some amino acids are coded by more than one codon
(iii) The sequence of triplet nitrogenous bases in DNA or mRNA corresponds to the amino acid sequence in the polypeptide chain.
Give suitable terms for the characteristics of ‘genetic code’ as per the above statements.CorrectIncorrectHint
(c)
-
Question 51 of 79
51. Question
The mutations that involve addition, deletion or substitution of a single base pair in a gene are referred to as
CorrectIncorrectHint
(a) : Most of the gene mutations involve a change in only a single nucleotide or nitrogen base of the cistron. These gene mutations are called point mutations e.g. sickle cell anemia in which polypeptide chain coding for haemoglobin contains valine instead of glutamic acid due to substitution of T by A in second position of triplet codon.
-
Question 52 of 79
52. Question
Sickle cell anemia results from a single base substitution in a gene, thus it is an example of
CorrectIncorrectHint
(a) : Most of the gene mutations involve a change in only a single nucleotide or nitrogen base of the cistron. These gene mutations are called point mutations e.g. sickle cell anemia in which polypeptide chain coding for haemoglobin contains valine instead of glutamic acid due to substitution of T by A in second position of triplet codon.
-
Question 53 of 79
53. Question
Select the incorrectly matched pair.
CorrectIncorrectHint
(d) : Polypeptide synthesis is signalled by two initiation codons – AUG (methionine) and GUG (valine). Polypeptide chain termination is signalled by three termination codons – UAA (ochre), UAG (amber) and UGA (opal). They do not specify any amino acid and are hence called non-sense codons. Anticodon loop is present on tRNA.
-
Question 54 of 79
54. Question
Identify the labels A, B, C and D in the given structure of tRNA and select the correct option.
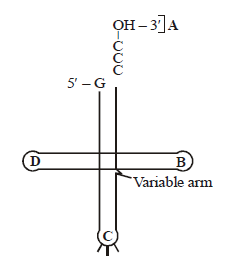 CorrectIncorrect
CorrectIncorrectHint
(b) In tRNA, there is a TWC loop which contains pseudouridine and ribothymidine. The loop is the site for attaching to ribosomes. Another loop, DHU loop contains dihydrouridine. It is a binding site for aminoacyl synthetase enzyme. tRNA molecules have unpaired (single-stranded) CCAOH sequence at the 3 end. This is called amino acid binding site because the amino acid becomes covalently attached to adenylic acid or A of CCA sequence during polypeptide synthesis. Anticodon loop is made up of three nitrogen bases for recognizing and attaching to the codon of mRNA.
-
Question 55 of 79
55. Question
The differences between mRNA and tRNA are that
(i) mRNA has more elaborated 3-dimensional structure due to extensive base-pairing
(ii) tRNA has more elaborated 3-dimensional structure due to extensive base-pairing
(iii) tRNA is usually smaller than mRNA
(iv) mRNA bears anticodon but tRNA has codons.CorrectIncorrectHint
(b) : mRNA is the longest RNA with maximum molecular weight but it is least abundant. IRNA is the smallest and coiled like a clover leaf with elaborated 3-dimensional structure. mRNA consists of 75-6000 bases while tRNA has 73-93 bases.
-
Question 56 of 79
56. Question
Amino acid acceptor end of tRNA lies at
CorrectIncorrectHint
(b)
-
Question 57 of 79
57. Question
Which RNA carries the amino acids from the amino acid pool to mRNA during protein synthesis?
CorrectIncorrectHint
(c) : Translation is the process of polymerisation by which the triplet base sequence of a MRNA guides the linking of a specific sequence of amino acids to form a polypeptides (protein) on ribosomes. Each \(\mathbb{R} N A\) is specific to an amino acid, as \(\mathbb{R} N A s\) are added to the sequence, amino acids are linked together by peptide bonds, eventually forming a protein that is later released by the \(\mathbb{R} N A\).
-
Question 58 of 79
58. Question
During translation, activated amino acids get linked to tRNA. This process is commonly called as
CorrectIncorrectHint
(d) : During translation, a \(\mathbb{R} N A\) is specifically linked to an amino acid, this process takes place under the direction of an enzyme, amino acyl \(\mathbb{R} N A\) synthetase that is extremely specific, i.e., recognises only one amino acid. tRNA complexed with amino acid is called as charged \(\mathbb{R} N \mathrm{NA}\). The process is referred to a charging or aminoacylation of \(\mathbb{R} N A\).
-
Question 59 of 79
59. Question
Refer to the given mRNA segment which can be translated completely into a polypeptide.
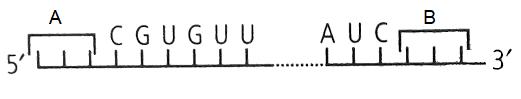
Which of the following codons may correspond with \(A\) and \(B\) ?
CorrectIncorrectHint
(a) : Polypeptide synthesis is signalled by two initiation codons – AUG (methionine) and GUG (valine). Polypeptide chain termination is signalled by three termination codons – UAA (ochre), UAG (amber) and UGA (opal). They do not specify any amino acid and are hence called non-sense codons. Anticodon loop is present on tRNA.
-
Question 60 of 79
60. Question
In a mRNA molecule, untranslated regions (UTRs) are present at
CorrectIncorrectHint
(c) : mRNA has some additional sequences that are not translated and are referred as untranslated regions (UTRs). The UTRs are present at both \(5^{\prime}\)-end (before start codon) and at \(3^{\prime}\)-end (after stop codon). They are required for efficient translation process.
-
Question 61 of 79
61. Question
UTRs are the untranslated regions present on
CorrectIncorrectHint
(c) : mRNA has some additional sequences that are not translated and are referred as untranslated regions (UTRs). The UTRs are present at both \(5^{\prime}\)-end (before start codon) and at \(3^{\prime}\)-end (after stop codon). They are required for efficient translation process.
-
Question 62 of 79
62. Question
Given below are the steps of protein synthesis. Arrange them in correct sequence and select the correct option.
(i) Codon-anticodon reaction between mRNA and aminoacyl tRNA complex
(ii) Attachment of mRNA and smaller sub-unit of ribosome
(iii) Charging or aminoacylation of tRNA
(iv) Attachment of larger sub-unit of ribosome to the mRNA-tRNA \({ }_{\text {Methyl }}\) complex
(v) Linking of adjacent amino acids.
(vi) Formation of polypeptide chain.CorrectIncorrectHint
(d)
-
Question 63 of 79
63. Question
Refer to the given figure.
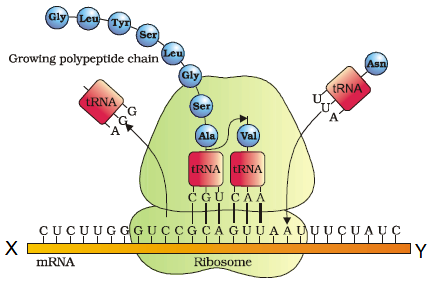
Select the option which identifies polarity X and Y and DNA sequence coding for serine P and the anticodon for the same amino acid (Q).\(
\begin{array}{|l|l|l|l|l|}
\hline & \mathbf{X} & \mathbf{Y} & \mathbf{P} & \mathbf{Q} \\
\hline \text { (a) } & 3^{\prime} & 5^{\prime} & T C A & U C A \\
\hline \text { (b) } & 5^{\prime} & 3^{\prime} & UUG & T C A \\
\hline \text { (c) } & 3^{\prime} & 5^{\prime} & U C A & T C A \\
\hline \text { (d) } & 5^{\prime} & 3^{\prime} & T C A & U C A \\
\hline
\end{array}
\)CorrectIncorrectHint
(d)
-
Question 64 of 79
64. Question
Which of the following statements is correct regarding ribosomes?
CorrectIncorrectHint
(b) : Ribosomes are called as protein factories. Each ribosome has two unequal parts, small and large. The larger subunit of ribosome has a groove for pushing out the newly formed polypeptide and protecting the same from cellular enzymes. The smaller subunit fits over the larger one like a cap but leaves a tunnel for mRNA. The two subunits come together only at the time of protein formation.
-
Question 65 of 79
65. Question
What would happen if in a gene encoding a polypeptide of 50 amino acids, \(25^{\text {th }}\) codon (UAU) is mutated to UAA?
CorrectIncorrectHint
(a) : UAA is a non-sense codon. It signals for polypeptide chain termination. Hence, only 24 amino acids chain will be formed.
-
Question 66 of 79
66. Question
Match column I with column II and select the correct option from the given codes.
\(
\begin{array}{|l|c|c|c|}
\hline & \text { Column I } & & \text { Column II } \\
\hline \text { A. } & \text { Translation } & \text { (i) } & \text { Aminoacyl tRNA synthetase } \\
\hline \text { B. } & \text { Transcription } & \text { (ii) } & \text { Okazaki fragments } \\
\hline \text { C. } & \text { DNA replication } & \text { (iii) } & \text { RNA polymerase } \\
\hline
\end{array}
\)CorrectIncorrectHint
(b)
-
Question 67 of 79
67. Question
Regulation of gene expression occurs at the level of
CorrectIncorrectHint
(d) : Regulation of gene expression can be exerted at four levels :
(i) transcriptional level during formation of primary transcript,
(ii) processing like splicing, terminal additions or modifications,
(iii) transport of mRNAs from nucleus to the cytoplasm and
(iv) translational level. -
Question 68 of 79
68. Question
During expression of an operon, RNA polymerase binds to
CorrectIncorrectHint
(d) : Promoter gene acts as an initiation signal which functions as recognition centre for RNA polymerase provided the operator gene is switched on. RNA polymerase is bound to the promoter gene. When the operator gene is functional, the polymerase moves over it and reaches the structural genes to perform transcription.
-
Question 69 of 79
69. Question
The sequence of structural genes in lac operon is
CorrectIncorrectHint
(d): The lac operon consists of three structural genes (lac z, lac y, lac a). Lac z codes for \(\beta\)-galactosidase which hydrolyses lactose to glucose and galactose. Lac y codes for lac permease, a membrane-bound protein constituent of the lactose transport system which increase permeability of the cell to \(\beta\)-galactosides. Lac a codes of thiogalactoside transacetylase, an enzyme of uncertain metabolic function.
-
Question 70 of 79
70. Question
Match column I with column II and select the correct option from the given codes.
\(
\begin{array}{|c|c|c|c|}
\hline & \text { Column I } & & \text { Column II } \\
\hline \text { A. } & \text { Operator site } & \text { (i) } & \begin{array}{l}
\text { Binding site for RNA } \\
\text { polymerase }
\end{array} \\
\hline \text { B. } & \text { Pro } & \text { (ii) } & \begin{array}{l}
\text { Binding site for repressor } \\
\text { molecule }
\end{array} \\
\hline \text { C. } & \text { Regulator gene } & \text { (iii) } & \text { Codes for protein / enzyme } \\
\hline \text { D. } & \text { Structural gene } & \text { (iv) } & \begin{array}{l}
\text { Codes for repressor } \\
\text { molecule }
\end{array} \\
\hline
\end{array}
\)CorrectIncorrectHint
(b)
-
Question 71 of 79
71. Question
Which of the following cannot act as inducer?
CorrectIncorrectHint
(d) : Lactose is the substrate for the enzyme \(\boldsymbol{\beta}\)-galactosidase and it regulates switching on and off of the operon and therefore, acts as inducer.
-
Question 72 of 79
72. Question
Inducible operon system usually occurs in (i) pathways. Nutrient molecules serve as (ii) to stimulate production of the enzymes necessary for their breakdown. Genes for inducible operon are usually switched (iii) and the repressor is synthesised in an (iv) form.
CorrectIncorrectHint
(c)
-
Question 73 of 79
73. Question
Repressible operon system is usually found in (i) pathways. The pathway’s end product serves as a (ii) to activate the repressor, turn off enzyme synthesis and prevent overproduction of the end product of the pathway. Genes for this operon are usually switched (iii) and the repressor is synthesised in an (iv) form.
CorrectIncorrectHint
(a)
-
Question 74 of 79
74. Question
Match column I with column II and select the correct option from the given codes.
\(
\begin{array}{|l|l|l|l|}
\hline & \text { Column I } & & \text { Column II } \\
\hline \text { A. } & \text { Griffith } & \text { (i) } & \text { Lac operon } \\
\hline \text { B. } & \text { Hershey and Chase } & \text { (ii) } & \begin{array}{l}
\text { Semi-conservative } \\
\text { DNA replication }
\end{array} \\
\hline
\end{array}
\)CorrectIncorrectHint
(a)
-
Question 75 of 79
75. Question
Human genome consists of approximately
CorrectIncorrectHint
(a) : Human genome has 3.1647 million nucleotide base pairs ( \(3 \times 10^9 \mathrm{bp}\) ). The average gene size is 3000 base pairs. The human genome consists of about 20,500 genes.
-
Question 76 of 79
76. Question
Estimated number of genes in human beings is
CorrectIncorrectHint
(c) : Human genome has 3.1647 million nucleotide base pairs ( \(3 \times 10^9 \mathrm{bp}\) ). The average gene size is 3000 base pairs. The human genome consists of about 20,500 genes.
-
Question 77 of 79
77. Question
Which of the following statements regarding ‘human genome’ is incorrect?
CorrectIncorrectHint
(d) : Repeated or repetitive sequences make up a large portion of human genome. Repetitive sequences are nucleotide sequences that are repeated many times, sometimes hundred to thousand times. They have no direct coding function but provide information as to chromosome structure, dynamics and evolution. Approximately 1 million copies of short 5-8 base pair repeated sequences are clustered around centromeres and near the ends
-
Question 78 of 79
78. Question
Select the correct option that correctly fill the blanks (i) – (iv).
I. Less than (i) of genome represents structural genes that code for proteins.
II. Chemical substance that binds with repressor and convert it into a non-DNA binding state is (ii).
III. In prokaryotes, during replication RNA primer is removed by (iii) whereas in eukaryotes it is removed by (iv).CorrectIncorrectHint
(c)
-
Question 79 of 79
79. Question
Arrange the various steps of DNA fingerprinting technique in the correct order.
(i) Separation of DNA fragments by electrophoresis.
(ii) Digestion of DNA by restriction endonucleases.
(iii) Hybridisation using labelled VNTR probe.
(iv) Isolation of DNA.
(v) Detection of hybridised DNA fragments by autoradiography.
(vi) Transferring the separated DNA fragments to nitrocellulose membrane.CorrectIncorrectHint
(a)